Description
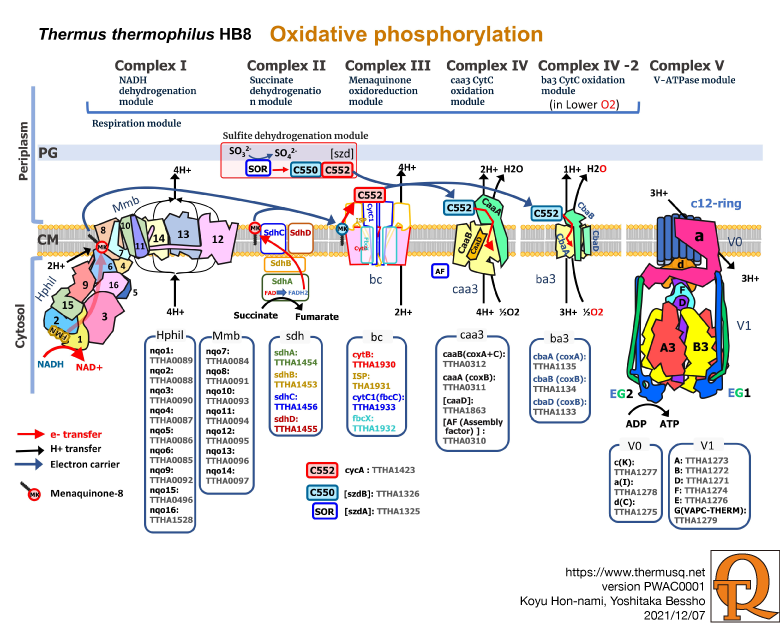
Thermus thermophilus HB8 oxidative phosphorylation complexes, cytochrome c552 as an electron career protein, and sulfite oxidase system with its (their) gene product(s). Among oxidative phosphorylation complexes, complexes I, IV-1, IV-2, and V, which have been structurally determined in this organism, are rimmed with black line. On the other hand, complexes II and III, structurally not determined yet, with color lines and in rough. T. thermophilus oxidative phosphorylation complexes are similar to those of mitochondria, except Complex IV branches into IV-1 and IV-2, probably due to adapting to oxygen tension of its habitat. This thermophile adopts menaquinone (MK), not but ubiquinone that is familiar to organism with mitochondria such as eukaryotes.
As in the oxidative phosphorylation, the electrons generated by the periplasmic sulfite oxidase system, via cytochrome c552, are used by Complex IVs for H+ translocation. PG and CM represent peptidoglycan and cytoplasmic membrane, respectively. NAD, FAD, FMN mean nicotinamide adenine dinucleotide, flavin adenine dinucleotide, flavin mononucleotide, respectively. SOR, C550, and C552 represent sulfite oxidoreductase, cyt c550, and cyt c552, respectively. In C552, C550, and MK, blue and red colors mean reduced and oxidized forms, respectively. Individual gene and gene product are also represented.
References
- Hon-Nami K, Hijikata A, Yura K, Bessho Y, (2023) Whole genome analyses for c-type cytochromes associated with respiratory chains in the extreme thermophile, Thermus thermophilus. J Gen Appl Microbiol, 69, 68–78.
- Hunsicker-Wang LM, Heine A, Chen Y, Luna EP, Todaro T, Zhang YM, Williams PA, McRee DE, Hirst J, Stout CD, et al., (2003) High-resolution structure of the soluble, respiratory-type Rieske protein from Thermus thermophilus: analysis and comparison. Biochemistry, 42, 7303–17.
- Kishikawa J-I, Nakanishi A, Furuta A, Kato T, Namba K, Tamakoshi M, Mitsuoka K, Yokoyama K, (2020) Mechanical inhibition of isolated Vo from V/A-ATPase for proton conductance. Elife, 9, .
- Zhou L, Sazanov LA, (2019) Structure and conformational plasticity of the intact Thermus thermophilus V/A-type ATPase. Science, 365, .
- Mather MW, Springer P, Fee JA, (1991) Cytochrome oxidase genes from Thermus thermophilus. Nucleotide sequence and analysis of the deduced primary structure of subunit IIc of cytochrome caa3. J Biol Chem, 266, 5025–35.
- Mather MW, Springer P, Hensel S, Buse G, Fee JA, (1993) Cytochrome oxidase genes from Thermus thermophilus. Nucleotide sequence of the fused gene and analysis of the deduced primary structures for subunits I and III of cytochrome caa3. J Biol Chem, 268, 5395–408.
- Lyons JA, Aragão D, Slattery O, Pisliakov AV, Soulimane T, Caffrey M, (2012) Structural insights into electron transfer in caa3-type cytochrome oxidase. Nature, 487, 514–8.
- Soulimane T, Than ME, Dewor M, Huber R, Buse G, (2000) Primary structure of a novel subunit in ba3-cytochrome oxidase from Thermus thermophilus. Protein Sci, 9, 2068–73.
- Soulimane T, Buse G, Bourenkov GP, Bartunik HD, Huber R, Than ME, (2000) Structure and mechanism of the aberrant ba(3)-cytochrome c oxidase from thermus thermophilus. EMBO J, 19, 1766–76.
- Robin S, Arese M, Forte E, Sarti P, Giuffrè A, Soulimane T, (2011) A sulfite respiration pathway from Thermus thermophilus and the key role of newly identified cytochrome c₅₅₀. J Bacteriol, 193, 3988–97.
- Mooser D, Maneg O, Corvey C, Steiner T, Malatesta F, Karas M, Soulimane T, Ludwig B, (2005) A four-subunit cytochrome bc(1) complex complements the respiratory chain of Thermus thermophilus. Biochim Biophys Acta, 1708, 262–74.
- Baradaran R, Berrisford JM, Minhas GS, Sazanov LA, (2013) Crystal structure of the entire respiratory complex I. Nature, 494, 443–8.
- Hinchliffe P, Sazanov LA, (2005) Organization of iron-sulfur clusters in respiratory complex I. Science, 309, 771–4.