Description
The purine nucleotide biosynthetic pathway is essentially common to most organisms. In terms of DNA replication and metabolism, the existence of a nucleotide biosynthetic pathway is necessary for the establishment of an organism, so it is important to investigate the establishment of this pathway in the early stages of life in order to consider the evolution of life. The purine nucleotide biosynthetic pathway has been investigated by Buchanan and colleagues in the 1950s and 1960s. By injecting pigeons with various isotope-labelled compounds and analyzing which atoms of the excreted uric acid were labelled. These researches determined which compound each atom of the purine ring was derived from. The purification of the enzymes involved in each reaction and the determination of the structures of the reaction intermediates were then carried out in sequence, and the reaction mechanism of the enzymes was also investigated.
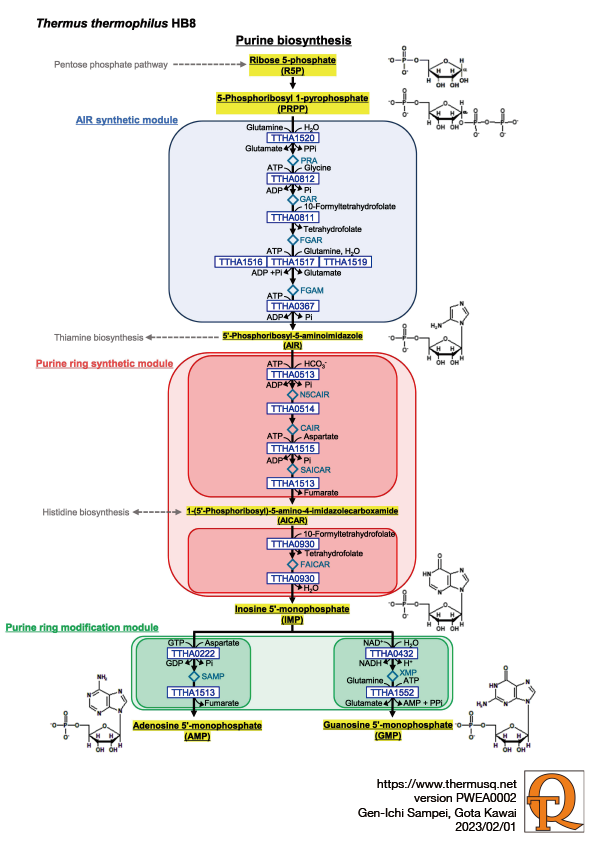
The purine nucleotide biosynthetic pathway consists of a total of 14 reactions, starting from 5-phosphoribosyl-1-pyrophosphate (PRPP), via inosine monophosphate (IMP), to adenosine monophosphate (AMP) and guanosine monophosphate (GMP). In E. coli, there are two enzymes (PurN and PurT) involved in the third reaction, using N10-formyltetrahydrofolate and formic acid as substrates, respectively. In contrast, in Thermus thermophilus, only PurN is involved in this step. For the sixth reaction, two enzymes, PurE (Class I) and PurK, are involved in some eubacteria (even in Thermus thermophilus), plants, yeasts and archaea, while in other eukaryotes and archaea, PurE (Class II) alone is known to be responsible for the reaction. In some species, PurL, involved in the fourth reaction, is divided into three subunits, PurQ, PurL and PurS (even in Thermus thermophilus), while GuaA, involved in the fourteenth reaction, is divided into two subunits, GuaA1 and GuaA2.
References
- HARTMAN SC, BUCHANAN JM, (1959) Nucleic acids, purines, pyrimidines (nucleotide synthesis). Annu Rev Biochem, 28, 365–410.
- HARTMAN SC, BUCHANAN JM, (1959) The biosynthesis of the purines. Ergeb Physiol, 50, 75–121.
- Buchanan JM, (1973) The Amidotransferases. , , 91–183.
- Sampei G, Mizobuchi K, (1989) The organization of the purL gene encoding 5'-phosphoribosylformylglycinamide amidotransferase of Escherichia coli. J Biol Chem, 264, 21230–21238.
- Kappock TJ, Ealick SE, Stubbe J, (2000) Modular evolution of the purine biosynthetic pathway. Curr Opin Chem Biol, 4, 567–572.
- Sampei G, Baba S, Kanagawa M, Yanai H, Ishii T, Kawai H, Fukai Y, Ebihara A, Nakagawa N, Kawai G, (2010) Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria. J Biochem, 148, 429–438.
- Kanagawa M, Baba S, Ebihara A, Shinkai A, Hirotsu K, Mega R, Kim K, Kuramitsu S, Sampei G, Kawai G, (2010) Structures of hypoxanthine-guanine phosphoribosyltransferase (TTHA0220) from Thermus thermophilus HB8. Acta Crystallogr Sect F Struct Biol Cryst Commun, 66, 893–898.
- Suzuki S, Yanai H, Kanagawa M, Tamura S, Watanabe Y, Fuse K, Baba S, Sampei G, Kawai G, (2012) Structure of N-formylglycinamide ribonucleotide amidotransferase II (PurL) from Thermus thermophilus HB8. Acta Crystallogr Sect F Struct Biol Cryst Commun, 68, 14–19.
- Sampei G, Kanagawa M, Baba S, Shimasaki T, Taka H, Mitsui S, Fujiwara S, Yanagida Y, Kusano M, Suzuki S, et al., (2013) Structures and reaction mechanisms of the two related enzymes, PurN and PurU. J Biochem, 154, 569–579.
- Kanagawa M, Baba S, Watanabe Y, Nakagawa N, Ebihara A, Kuramitsu S, Yokoyama S, Sampei G, Kawai G, (2015) Crystal structures and ligand binding of PurM proteins from Thermus thermophilus and Geobacillus kaustophilus. J Biochem, mvv107, .
- Watanabe Y, Yanai H, Kanagawa M, Suzuki S, Tamura S, Okada K, Baba S, Kumasaka T, Agari Y, Chen L, et al., (2016) Crystal structures of a subunit of the formyl-glycinamide ribonucleotide amidotransferase, PurS, from Thermus thermophilus, Sulfolobus tokodaii and Methanocaldococcus jannaschii. Acta Crystallogr Sect F Struct Biol Commun, 72, 627–635.
- Yamamoto N, Sampei G, Kawai G, (2022) Free-Energy Profile Analysis of the Catalytic Reaction of Glycinamide Ribonucleotide Synthetase. Life, 12, 281.
- Sampei G, Ishii H, Taka H, Kawai G, (2023) Convergent evolution of nitrogen-adding enzymes in the purine nucleotide biosynthetic pathway, based on structural analysis of adenylosuccinate synthetase (PurA). J Gen Appl Microbiol, 69, 109–116.